Machine Learning of Force Fields for Molecular Dynamics Simulations of Proteins at DFT Accuracy
ICLR 2024 GEM Workshop May 2024

Protein binding affinity prediction under multiple substitutions based on eGNNs with residue and atomic graphs and language model information: eGRAL
ICLR 2024 GEM Workshop May 2024
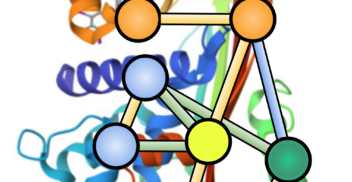
Exploring Genomic Language Models on Protein Downstream Tasks
LLMs4Bio AAAI 2024 | MLGenX ICLR 2024 May 2024
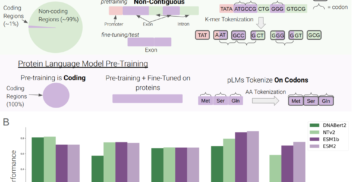
Advancing DNA Language Models: The Genomics Long-Range Benchmark
LLMs4Bio AAAI 2024 | MLGenX ICLR 2024 May 2024

SegmentNT: annotating the genome at single-nucleotide resolution with DNA foundation models
Mar 2024

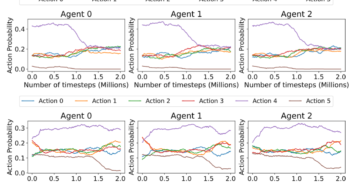
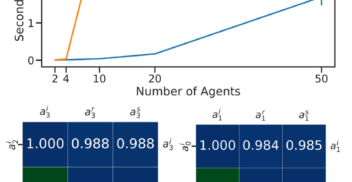
How much can change in a year? Revisiting Evaluation in Multi-Agent Reinforcement Learning
AAAI workshop Mar 2024
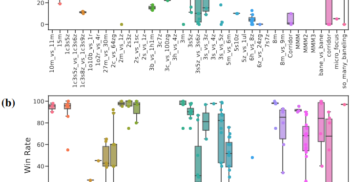
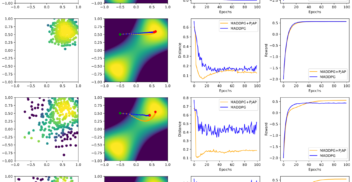
On Diagnostics for Understanding Agent Training Behaviour in Cooperative MARL
AAAI workshop Mar 2024