Please read our extensive Privacy policy here. You can also read our Privacy Notice and our Cookie Notice
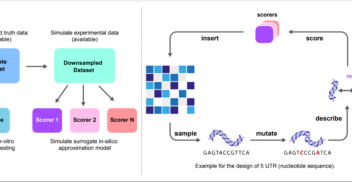
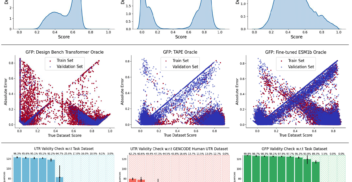
Overconfident Oracles: Limitations of In Silico Sequence Design Benchmarking
ICML 2024 workshop Jul 2024
Generative Model for Small Molecules with Latent Space RL Fine-Tuning to Protein Targets
ICML 2024 workshop Jul 2024
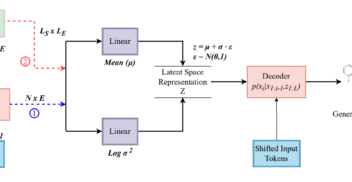
Likelihood-based fine-tuning of protein language models for few-shot fitness prediction and design
ICML 2024 workshop Jul 2024
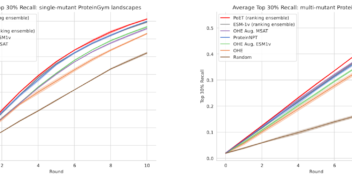
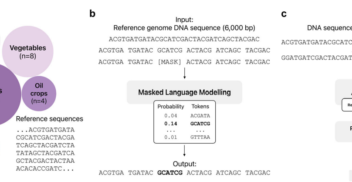
A large language foundational model for edible plant genomes
Nature Communications Biology 2024 Jul 2024
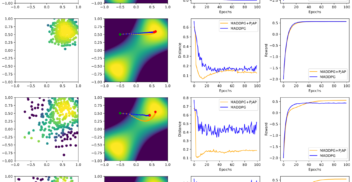
Machine Learning of Force Fields for Molecular Dynamics Simulations of Proteins at DFT Accuracy
ICLR 2024 GEM Workshop May 2024