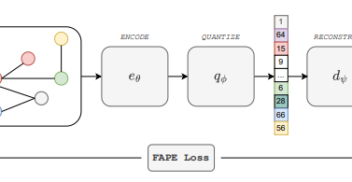
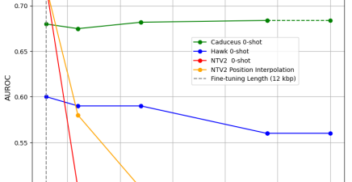
De novo peptide sequencing with InstaNovo: Accurate, database-free peptide identification for large scale proteomics experiments
Nature Machine Intelligence Mar 2025
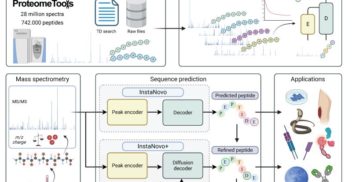
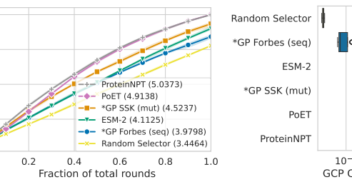
Bayesian Optimisation for Protein Sequence Design: Gaussian Processes with Zero-Shot Protein Language Model Prior Mean
NeurIPS 2024 workshop Dec 2024
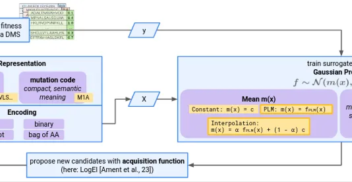
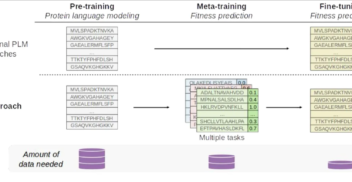
Bayesian Optimisation for Protein Sequence Design: Back to Basics with Gaussian Process Surrogates
NeurIPS 2024 workshop Dec 2024


![Free energy surface of unseen alanine-dipeptide Comparison of the samples obtained by running ground truth MD and boostMD. The free energy of the Ramachandran plot, is directly related to the marginalized Boltzmann distribution exp [−F(ϕ, ψ)/kBT]. The reference model is evaluated every 10 steps. Both simulations are run for 5 ns (5 × 106 steps).](https://www.instadeep.com/wp-content/uploads/2024/12/image-7-352x182.png)