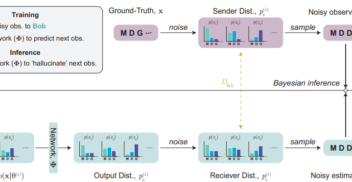
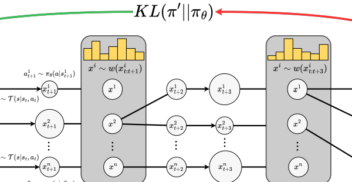
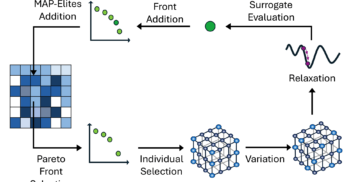
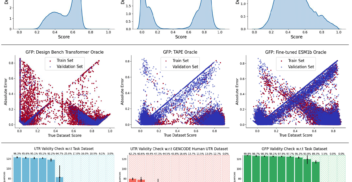
On 1st October 2024, over 100 members of the biotech community – including scientists, researchers, engineers and journalists – gathered at London’s tech hub, CodeNode, for...

AI Day 2024: InstaDeep Showcases Innovations in Biology and AI as...
on Oct 09, 2024 | 10:40am