As AI continues to reshape our understanding of the biological landscape, DNA, RNA, and protein sequence models are rapidly emerging—each promising to be faster, more capable, and more accurate than the last.
While genomic data is being generated at an astonishing pace, making sense of it remains a challenge. A key barrier is usability. Most lab scientists don’t have the coding background required to work with today’s models. At the same time, current AI developments in genomics tend to be narrow specialists. They excel at one task but struggle to generalise, often needing extensive reprogramming and fine-tuning to adapt to new problems. This limited efficiency slows the pace of discovery, but we believe Chat Nucleotide Transformer (ChatNT) has the potential to change that.
What is ChatNT?
As seen in Nature Machine Intelligence, ChatNT is the latest addition to the Nucleotide Transformer (NT) family: a generative AI model that unites biological sequences and natural language in a single conversational interface.
Inspired by large vision language models (vLLMs) like GPT-4o or Llama4—systems that process both text and image—our researchers reimagined genomics prediction tasks as text-to-text problems, designing a multimodal DNA-meets-language agent that enables seamless interaction.
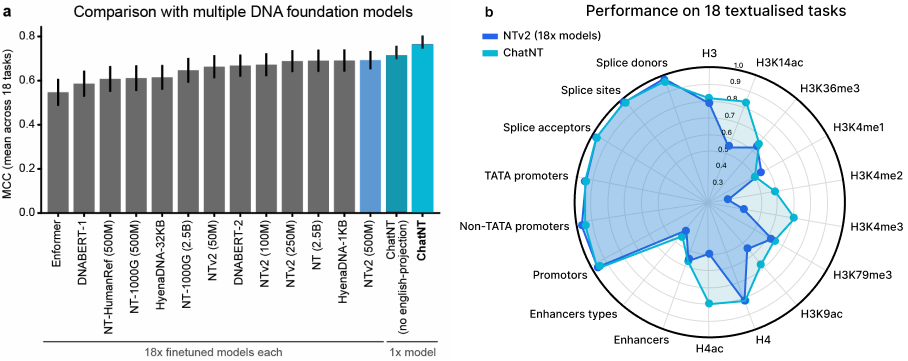
ChatNT is a generalist genomics AI, a unified model that can interpret multiple biological sequences and handle dozens of tasks. To evaluate performance, we tested ChatNT on the publicly available Nucleotide Transformer (NT) benchmark, a framework introduced by InstaDeep in earlier work that assesses biological sequence models on a set of 18 genomics tasks. ChatNT achieved state-of-the-art results with a Matthews Correlation Coefficient (MCC) of 0.77, outperforming the best prior model (Nucleotide Transformer V2), which required separate models per task.
💡MCC is a measure of how well the model’s predictions match the real biological data. An MCC of 1 means perfect prediction, 0 means random guessing, and -1 means perfectly incorrect prediction (i.e. every outcome is the opposite of the true label).
How it works
ChatNT was trained on a new, large-scale dataset spanning 27 biological functions and species. The dataset includes over 600 million DNA tokens and 273 million English tokens—providing unprecedented training volume for task-driven genomics models.
The model processes biological sequences in three key steps:
- DNA Encoder (NTv2):
Breaks DNA into six nucleotide chunks and converts them into dense numerical vectors that represent meaningful biological patterns. - Projection & Translation:
These vectors are reshaped and filtered through a neural network and a perceiver resampler that selects the most relevant information based on the query. - English Decoder (Vicuna-7B):
The refined DNA information is embedded into the prompt and passed to a frozen language model that interprets it in context and returns a response in plain English.
What can it do?
To evaluate ChatNT’s real-world applicability, we tested the model across a range of use cases designed to position it as a highly efficient assistant for genomics research.
Measuring model confidence
One of the biggest concerns with language models is hallucination—when a model generates incorrect information. ChatNT addresses this by learning to estimate its own confidence, making it more trustworthy and suitable to sensitive research applications.
To gauge this, our researchers introduced a perplexity-based scoring method, tailored for binary classification tasks. Rather than simply returning a “yes” or “no” answer, ChatNT evaluates how surprised it is by each possible response. It calculates a perplexity score for both answers, and our researchers convert these into probabilities. To ensure these estimates are reliable, we applied Platt scaling (a calibration technique that aligns predicted probabilities with actual performance) and found that ChatNT is well-calibrated in both low and high-confidence zones.
💡Lower perplexity = higher confidence. The model finds outputs more predictable based on its internal understanding.
While not yet integrated into the live ChatNT interface, this scoring method is general enough to extend beyond genomics, presenting a potentially promising approach for building more trustworthy conversational agents.
Model interpretability
Reinforcing this confidence is ChatNT’s interpretability—its ability to show scientists why it predicts something.
Using gradient-based attribution, our researchers traced how different parts of a DNA input influenced ChatNT’s natural language output across a variety of sequences and tasks. The results revealed that ChatNT’s predictions are often based on biologically meaningful motifs.
In splice site prediction, for example, the model consistently attributed high importance to GT and AG dinucleotides—classic indicators of donor and acceptor sites. Similarly, in promoter prediction, ChatNT highlighted TATA boxes, which are well-known markers for transcription initiation.
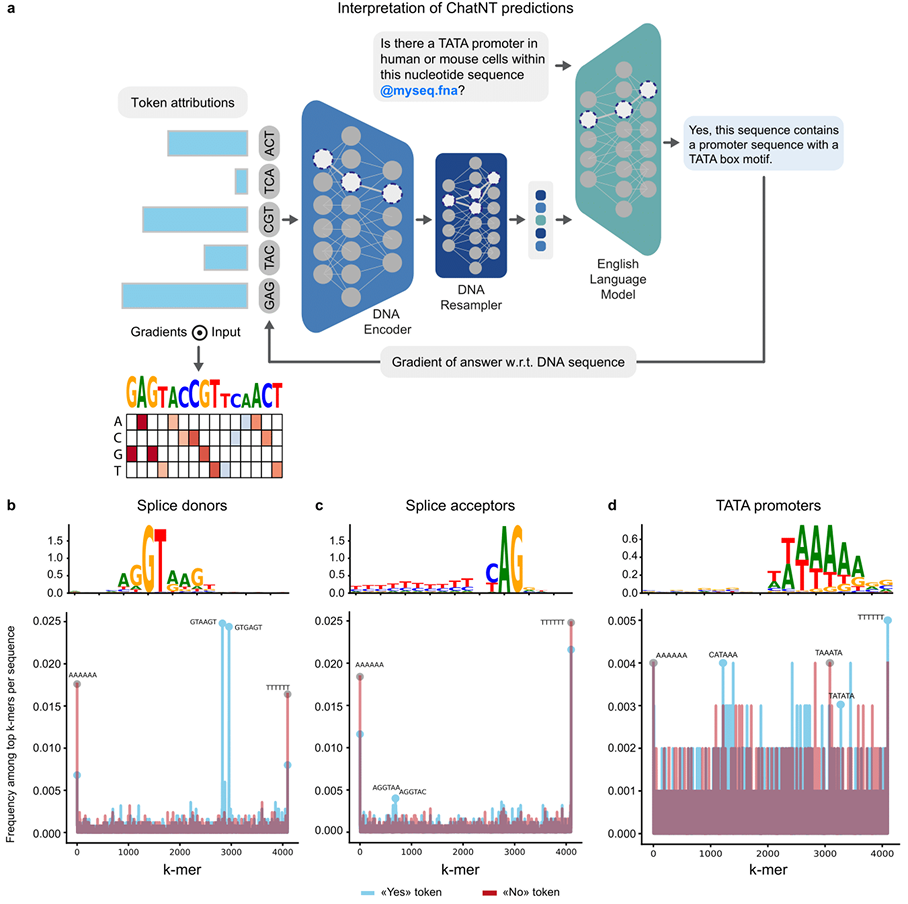
What’s interesting is that ChatNT achieves this as a single, unified model. The consistency across domains suggests it has learned key features of genomic grammar and applies this understanding through deeply grounded biological reasoning.
It even demonstrated the ability to link English words to biological patterns. When prompted about a “promoter” ChatNT didn’t simply parrot a memorised response, it identified real sequence motifs associated with the enhancer function.
Genomics
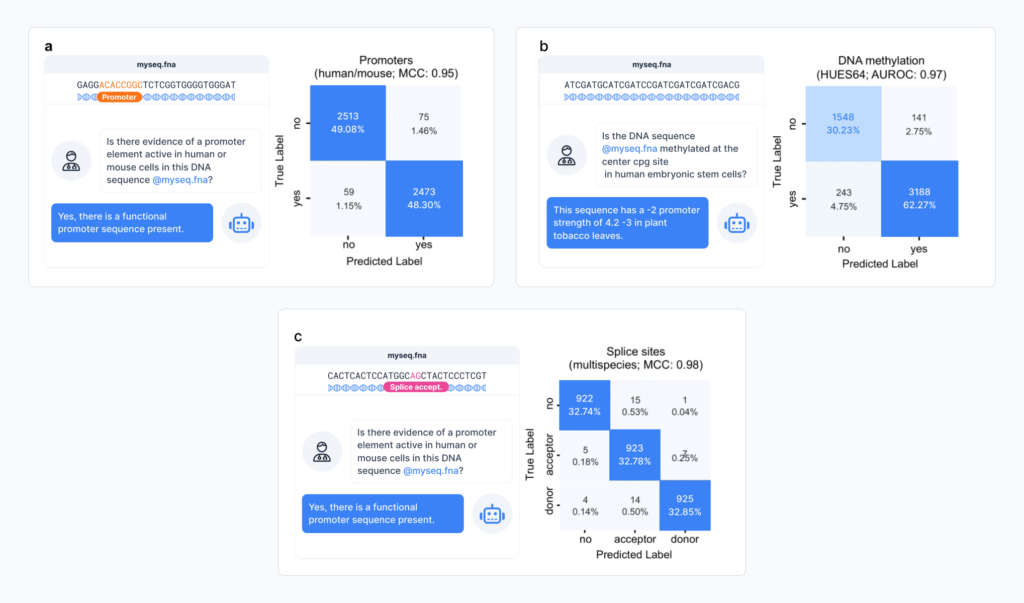
ChatNT has strong multi-task capabilities across core DNA-based tasks, excelling at both classification and regression. It can predict chromatin and histone features, identify key regulatory regions like promoters and enhancers, and locate splice sites, all from raw sequence input guided by a natural language prompt.
For instance, it achieved an MCC of 0.95 for human promoter prediction, and a 0.98 for splice site detection. These results are on par with state-of-the-art specialised models, yet ChatNT delivers them all through a single conversational interface.
While performance does vary across species and domains, ChatNT surpassed previous models in detecting human enhancer types. It did, however, show weaker performance on plant enhancer prediction compared to AgroNT, a model pre-trained on 48 plant genomes. This reflects ChatNT’s strength as a generalist and also points to future opportunities for targeted improvement.
Proteomics
ChatNT generalises beyond DNA to handle RNA and protein tasks using the same conversational approach.
To test this, our researchers included six regression tasks in the instructions dataset: three RNA and three protein-related. We asked the model to generate numeric outputs from sequence-based prompts, as shown in Figure 4.
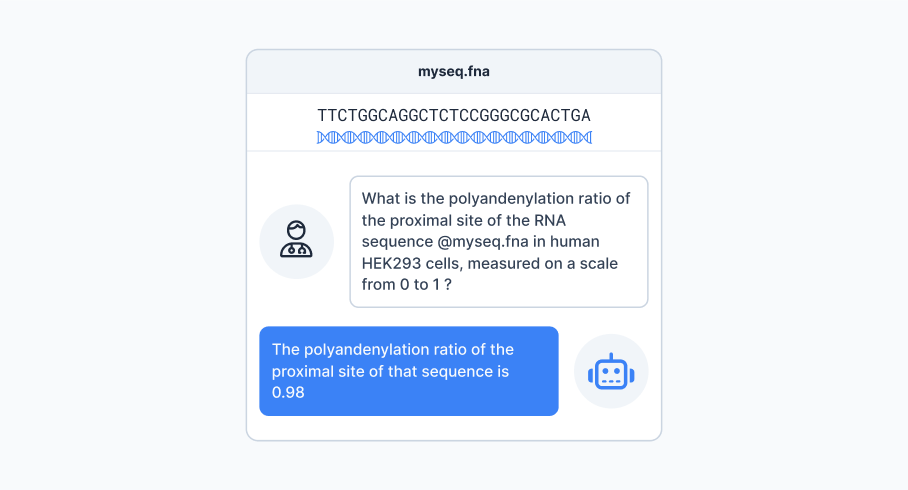
Despite being generalist, ChatNT achieved competitive performance across all six tasks. It outperformed state-of-the-art models like APARENT2 on proximal polyadenylation prediction— achieving a Pearson Correlation Coefficient (PCC) score of 0.91 vs 0.90—and ESM2 on protein melting point prediction (PCC: 0.89 vs 0.85). It also produced highly correlated outputs for other protein features like fluorescence and stability.
💡PCC measures how well predicted values correlate with true values (1 = perfect match).
ChatNT also delivered high correlation scores for other tasks like protein fluorescence and stability, all while using a DNA-centric encoder (NTv2), demonstrating its architectural flexibility.
What’s next?
ChatNT represents a fundamental shift in how we approach genomics. Looking ahead, future iterations could evolve far beyond phenotype prediction and numeric output, extending into broader bioinformatics workflows.
The model’s architecture is designed for scalability. Future efforts to expand the number and diversity of training tasks could potentially enhance ChatNT’s zero-shot generalisation, solving entirely new tasks without the need for retraining.
Multimodality remains a key area for growth. Incorporating RNA and protein-specific encoders, supported by a shared positional tagging system, would enable unified inputs. Upgrading the English decoder and expanding the sequence context window would further enhance ChatNT’s capabilities and range.
As it stands, ChatNT is our first proof-of-concept for a multimodal conversational agent capable of solving advanced and biologically relevant tasks. In a field that has long awaited intuitive high-performance tools, ChatNT lays the foundation for future generalist AI systems, marking an important step toward multi-purpose AI for biology and medicine.
Ready to explore ChatNT? Download the paper and access our open-source code, today! Now available on GitHub and HuggingFace.
Disclaimer: All claims made are supported by our research paper: A multimodal conversational agent for DNA, RNA, and protein tasks unless explicitly cited otherwise.
